Showing posts with label programming. Show all posts
Showing posts with label programming. Show all posts
Wednesday, April 6, 2016
Tuesday, April 5, 2016
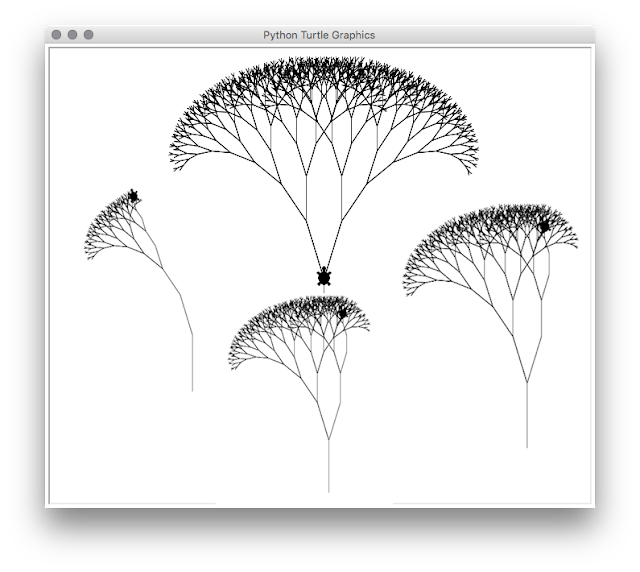
Fractal Tree using Python with Turtle Module
Here is "minus one" nearly symmetrical fractal tree
.
import turtle
import numpy
#buat pola di sini
#kura-kura menghadap ke atas
turtle.shape("turtle")
turtle.left(90)
lv = 7
l = 100
s = 17
turtle.penup()
turtle.backward(l)
turtle.pendown()
turtle.forward(l)
def maju(l,level):
l = 3./4.*l
turtle.left(s)
turtle.forward(l)
if level<lv:
level +=1
maju(l,level)
turtle.backward(l)
turtle.right(2*s)
turtle.forward(l)
if level<lv:
maju(l,level)
turtle.backward(l)
turtle.left(s)
level -=1
maju(l,2)
#agar gambar tak langsung hilang
turtle.exitonclick()
if we want symmetrical result, just move
level +=1
syntax
to the place before first if
Some Mistake Often Provide Beautiful Result.
:)
Planned to coding tree branch of tree, fractal mode, using turtle module on python. Has some trouble on backward rule. It become flower, by definition, it's still tree, :)
.
Planned to coding tree branch of tree, fractal mode, using turtle module on python. Has some trouble on backward rule. It become flower, by definition, it's still tree, :)
import turtle
#buat pola di sini
#kura-kura menghadap ke atas
turtle.shape("turtle")
turtle.left(90)
s = 37
lv = 7
l = 100
turtle.forward(l)
def mundur(l,level):
l = 4./3.*l
turtle.backward(l)
turtle.right(3*s)
maju(l,level)
def maju(l,level):
l = 3./4.*l
turtle.left(s)
turtle.forward(l)
level +=1
if level<=lv:
maju(l,level)
else:
mundur(l,level)
maju(l,2)
#agar gambar tak langsung hilang
turtle.exitonclick()
Wednesday, March 30, 2016
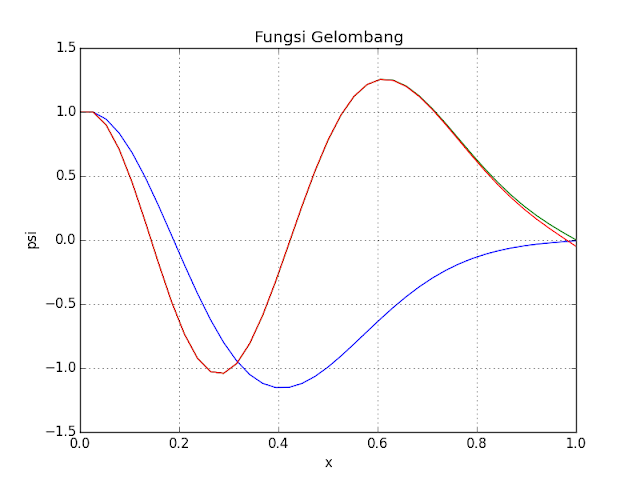
Even Wave Equation on Infinite-depth Potential Well (with Some Variation Potential in Between) using Shooting Method,
It's from previous code, I use it to simulate even function.
.
Here's some result with different potential V
from pylab import *
figure(3)
n = 39
psi0= zeros(n) #
psi = psi0 #
x = linspace(0,1,n)
V = zeros(n)
#V = 39*pow(x,2)
for i in arange(n):
if i<n/2.:
V[i] = 73.
else:
V[i] = 0.
psi0[0] = 1.
psi0[1] = psi0[0] #for odd function, use another value,
#but psi0[0] must be zero
t = 0
dx = 1./n
E = 1.
dE = .1
err = .05
while t< 771:
#k = 2*dx*dx*(E-V)
for i in range (1,n-1):
k = 2*dx*dx*(E-V[i])
psi[i+1] = 2*psi0[i]-psi0[i-1]-k*psi0[i]
psi0 = psi
if abs(psi[n-1])<err:
print E
plot(x,psi)
t += 1
E += dE
xlabel('x')
ylabel('psi')
title('Fungsi Gelombang')
grid(True)
savefig("shooting.png")
figure(2)
plot(x,V)
xlabel('x')
ylabel('V')
title('Potensial')
grid(True)
show()
Here's some result with different potential V
Monday, March 28, 2016
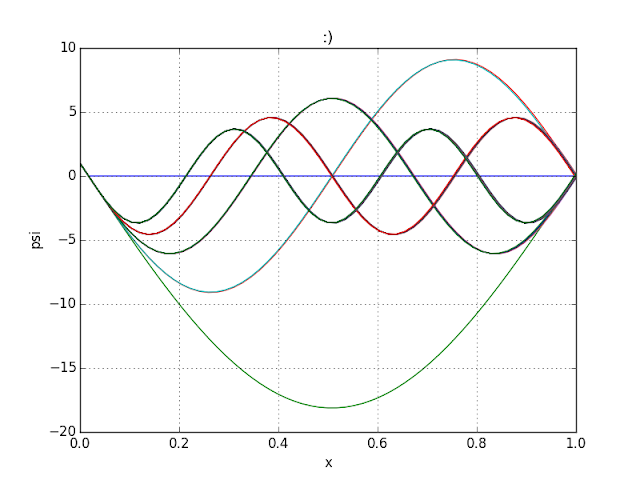
Energy Quantization on Potential Well; Shooting Method, :) .
Using Pylab module
Unlike the code before, only chosen energy with psi=0 on both end is plotted.
.
Unlike the code before, only chosen energy with psi=0 on both end is plotted.
from pylab import *
n = 59
psi0= zeros(n)
psi = psi0
x = linspace(0,1,n)
psi0[0] = 1.
plot(x,psi)
t = 0
dx = 1./16.
E = 0.
dE = .2
V = 0.
while t< 1337:
t += 1
E += .01
k = 2*dx*dx*(E-V)
for i in range (1,n-1):
psi[i+1] = 2*psi0[i]-psi0[i-1]-k*psi0[i]
#psi[2:] = 2*psi0[1:-1]-psi0[:-2]-k*psi0[1:-1]
psi0 = psi
#print psi[n-1]
if abs(psi[n-1])<=dE:
#pass
print E
plot(x,psi)
xlabel('x')
ylabel('psi')
title(':)')
grid(True)
savefig("els.png")
show()
Thursday, March 24, 2016
Shooting Method on Potential Well
It's the base code I wrote using Python, still need improvement to get energy level or even what energy allowed in the system.
.
from pylab import *
n = 19
psi0= zeros(n)
psi = psi0
x = linspace(0,1,n)
psi0[0] = 1.
plot(x,psi)
t = 0
dx = 1./8.
E = .1
V = 0.
while t< 27:
t += 1
E += .2
k = 2*dx*dx*(E-V)
for i in range (1,n-1):
psi[i+1] = 2*psi0[i]-psi0[i-1]-k*psi0[i]
psi0 = psi
plot(x,psi)
xlabel('x')
ylabel('psi')
title(':)')
grid(True)
savefig("els.png")
show()
Sunday, March 6, 2016
DLA Cluster in Python
It's using random parameter, so if it'll show the different result every time it's executed.
.
"""
Cluster
"""
import numpy as np #untuk operasi array
import matplotlib.pyplot as plt #untuk gambar grafik
import matplotlib.animation as animation #untuk menggerakkan grafik
fig, ax = plt.subplots()
plt.ylim(0,40)
plt.xlim(0,40)
#variabel
n = 39
x0 = 19
y0 = 19
a = np.zeros((n,n))
a[x0,y0] = 1
#print a
rx = []
ry = []
#membuat garis/kurva dengan sumbu-x adalah x, sumbu-y adalah y
line, = ax.plot(x0, y0, 'o')
x0 = np.random.randint(n)
y0 = np.random.randint(n)
x = x0
y = y0
def animate(i):
global line
global x0,y0,rx,ry,n
#menentukan seed baru
x = x0 + np.random.randint(-1,2)
y = y0 + np.random.randint(-1,2)
#print 'x = ', x, ' y = ', y
#apakah keluar batas?
#apakah menyentuh cluster utama?
if (x>(n-2)) or (y>(n-2)) or (x<1) or (y<1) or\
(a[x,y-1] + a[x,y] + a[x,y+1] + a[x-1,y-1] + a[x-1,y] + a[x-1,y+1] +\
a[x+1,y-1] + a[x+1,y] + a[x+1,y+1]) >= 1:
#renew()
#update subcluster menjadi bagian dari cluster (0 ke 1)
a[x,y] = 1
for i in np.arange(len(rx)):
a[rx[i],ry[i]] = 1
#print a
ok = 0
while ok==0:
x = np.random.randint(n)
y = np.random.randint(n)
#cek
if (a[x,y]==0) and (x>1) and (y>1) and (x<(n-2))and (y<(n-2)):
ok = 1
ok = 0
#kosongkan
#print 'rx', rx
rx = []
ry = []
#print 'rx',rx
#print 'xnew = ', x, ' y = ', y
#setelah mendapatkan seed baru, rekam titiknya
rx.append(x)
ry.append(y)
x0 = x
y0 = y
line, = ax.plot(x,y,'o')
return line,
ani = animation.FuncAnimation(fig, animate, frames=777, interval=100, blit=False)
ani.save('clusterDLA.mp4',bitrate=1024)
#plt.show()
Friday, November 27, 2015
3D (Polar/Cylindrical Coordinate) Animation of 2D Diffusion Equation using Python, Scipy, and Matplotlib
Yup, that same code but in polar coordinate.
I use nabla operator for cylindrical coordinate but ditch the z component.
So, what's the z-axis for? It's represent the u value, in this case, temperature, as function of r and phi (I know I should use rho, but, ...)
.
I use nabla operator for cylindrical coordinate but ditch the z component.
So, what's the z-axis for? It's represent the u value, in this case, temperature, as function of r and phi (I know I should use rho, but, ...)
import scipy as sp
from mpl_toolkits.mplot3d import Axes3D
from matplotlib import cm
from matplotlib.ticker import LinearLocator, FormatStrFormatter
import matplotlib.pyplot as plt
import mpl_toolkits.mplot3d.axes3d as p3
import matplotlib.animation as animation
#dr = .1
#dp = .1
#nr = int(1/dr)
#np = int(2*sp.pi/dp)
nr = 10
np = 10
r = sp.linspace(0.,1.,nr)
p = sp.linspace(0.,2*sp.pi,np)
dr = r[1]-r[0]
dp = p[1]-p[0]
a = .5
tmax = 100
t = 0.
dr2 = dr**2
dp2 = dp**2
dt = dr2 * dp2 / (2 * a * (dr2 + dp2) )
dt /=10.
print 'dr = ',dr
print 'dp = ',dp
print 'dt = ',dt
ut = sp.zeros([nr,np])
u0 = sp.zeros([nr,np])
ur = sp.zeros([nr,np])
ur2 = sp.zeros([nr,np])
#initial
for i in range(nr):
for j in range(np):
if ((i>4)&(i<6)):
u0[i,j] = 1.
#print u0
def hitung_ut(ut,u0):
for i in sp.arange (len(r)):
if r[i]!= 0.:
ur[i,:] = u0[i,:]/r[i]
ur2[i,:] = u0[i,:]/(r[i]**2)
ut[1:-1, 1:-1] = u0[1:-1, 1:-1] + a*dt*(
(ur[1:-1, 1:-1] - ur[:-2, 1:-1])/dr+
(u0[2:, 1:-1] - 2*u0[1:-1, 1:-1] + u0[:-2,1:-1])/dr2+
(ur2[1:-1, 2:] - 2*ur2[1:-1, 1:-1] + ur2[1:-1, :-2])/dp2)
#calculate the edge
ut[1:-1, 0] = u0[1:-1, 0] + a*dt*(
(ur[1:-1, 0] - ur[:-2, 0])/dr+
(u0[2:, 0] - 2*u0[1:-1, 0] + u0[:-2, 0])/dr2+
(ur2[1:-1, 1] - 2*ur2[1:-1, 0] + ur2[1:-1, np-1])/dp2)
ut[1:-1, np-1] = u0[1:-1, np-1] + a*dt*(
(ur[1:-1, np-1] - ur[:-2, np-1])/dr+
(u0[2:, np-1] - 2*u0[1:-1, np-1] + u0[:-2,np-1])/dr2+
(ur2[1:-1, 0] - 2*ur2[1:-1, np-1] + ur2[1:-1, np-2])/dp2)
#hitung_ut(ut,u0)
#print ut
def data_gen(framenumber, Z ,surf):
global ut
global u0
global t
hitung_ut(ut,u0)
u0[:] = ut[:]
Z = u0
t += 1
print t
ax.clear()
plotset()
surf = ax.plot_surface(X, Y, Z, rstride=1, cstride=1, cmap=cm.coolwarm,
linewidth=0, antialiased=False, alpha=0.7)
return surf,
fig = plt.figure()
#ax = fig.gca(projection='3d')
ax = fig.add_subplot(111, projection='3d')
P,R = sp.meshgrid(p,r)
X,Y = R*sp.cos(P),R*sp.sin(P)
Z = u0
print len(R), len(P)
def plotset():
ax.set_xlim3d(-1., 1.)
ax.set_ylim3d(-1., 1.)
ax.set_zlim3d(-1.,1.)
ax.set_autoscalez_on(False)
ax.zaxis.set_major_locator(LinearLocator(10))
ax.zaxis.set_major_formatter(FormatStrFormatter('%.02f'))
cset = ax.contour(X, Y, Z, zdir='x', offset=-1. , cmap=cm.coolwarm)
cset = ax.contour(X, Y, Z, zdir='y', offset=1. , cmap=cm.coolwarm)
cset = ax.contour(X, Y, Z, zdir='z', offset=-1., cmap=cm.coolwarm)
plotset()
surf = ax.plot_surface(X, Y, Z, rstride=1, cstride=1, cmap=cm.coolwarm,
linewidth=0, antialiased=False, alpha=0.7)
fig.colorbar(surf, shrink=0.5, aspect=5)
ani = animation.FuncAnimation(fig, data_gen, fargs=(Z, surf),frames=4096, interval=4, blit=False)
#ani.save('2dDiffusionfRadialf1024b512.mp4', bitrate=1024)
plt.show()
 |
| 100x100 size |
Thursday, November 26, 2015
The Wrong Code Will often Provide Beautiful Result, :)
It means to compute 2d diffusion equation just like previous post in polar/cylindrical coordinate, and all went to wrong direction, :)
Still trying to understand matplotlib mplot3d behavior
.
Still trying to understand matplotlib mplot3d behavior
import scipy as sp
from mpl_toolkits.mplot3d import Axes3D
from matplotlib import cm
from matplotlib.ticker import LinearLocator, FormatStrFormatter
import matplotlib.pyplot as plt
import mpl_toolkits.mplot3d.axes3d as p3
import matplotlib.animation as animation
#dr = .1
#dp = .1
#nr = int(1/dr)
#np = int(2*sp.pi/dp)
nr = 10
np = 10
dr = 1./nr
dp = 2*sp.pi/np
a = .5
tmax = 100
t = 0.
dr2 = dr**2
dp2 = dp**2
dt = dr2 * dp2 / (2 * a * (dr2 + dp2) )
dt /=10.
print dt
ut = sp.zeros([nr,np])
u0 = sp.zeros([nr,np])
ur = sp.zeros([nr,np])
ur2 = sp.zeros([nr,np])
r = sp.arange(0.,1.,dr)
p = sp.arange(0.,2*sp.pi,dp)
#initial
for i in range(nr):
for j in range(np):
if ( (i>(2*nr/5.)) & (i<(3.*nr/3.)) ):
u0[i,j] = 1.
#print u0
def hitung_ut(ut,u0):
for i in sp.arange (len(r)):
if r[i]!= 0.:
ur[i,:] = u0[i,:]/r[i]
ur2[i,:] = u0[i,:]/(r[i]**2)
ut[1:-1, 1:-1] = u0[1:-1, 1:-1] + a*dt*(
(ur[1:-1, 1:-1] - ur[:-2, 1:-1])/dr+
(u0[2:, 1:-1] - 2*u0[1:-1, 1:-1] + u0[:-2,1:-1])/dr2+
(ur2[1:-1, 2:] - 2*ur2[1:-1, 1:-1] + ur2[1:-1, :-2])/dp2)
#hitung_ut(ut,u0)
#print ut
def data_gen(framenumber, Z ,surf):
global ut
global u0
hitung_ut(ut,u0)
u0[:] = ut[:]
Z = u0
ax.clear()
plotset()
surf = ax.plot_surface(X, Y, Z, rstride=1, cstride=1, cmap=cm.coolwarm,
linewidth=0, antialiased=False, alpha=0.7)
return surf,
fig = plt.figure()
#ax = fig.gca(projection='3d')
ax = fig.add_subplot(111, projection='3d')
R = sp.arange(0,1,dr)
P = sp.arange(0,2*sp.pi,dp)
R,P = sp.meshgrid(R,P)
X,Y = R*sp.cos(P),R*sp.sin(P)
Z = u0
print len(R), len(P)
def plotset():
ax.set_xlim3d(-1., 1.)
ax.set_ylim3d(-1., 1.)
ax.set_zlim3d(-1.,1.)
ax.set_autoscalez_on(False)
ax.zaxis.set_major_locator(LinearLocator(10))
ax.zaxis.set_major_formatter(FormatStrFormatter('%.02f'))
cset = ax.contour(X, Y, Z, zdir='x', offset=0. , cmap=cm.coolwarm)
cset = ax.contour(X, Y, Z, zdir='y', offset=1. , cmap=cm.coolwarm)
cset = ax.contour(X, Y, Z, zdir='z', offset=-1., cmap=cm.coolwarm)
plotset()
surf = ax.plot_surface(X, Y, Z, rstride=1, cstride=1, cmap=cm.coolwarm,
linewidth=0, antialiased=False, alpha=0.7)
fig.colorbar(surf, shrink=0.5, aspect=5)
ani = animation.FuncAnimation(fig, data_gen, fargs=(Z, surf),frames=500, interval=30, blit=False)
#ani.save('2dDiffusionf500b512.mp4', bitrate=512)
plt.show()
Wednesday, November 25, 2015
3D Animation of 2D Diffusion Equation using Python, Scipy, and Matplotlib
I wrote the code on OS X El Capitan, use a small mesh-grid. Basically it's same code like the previous post.
I use surface plot mode for the graphic output and animate it.
Because my Macbook Air is suffered from running laborious code, I save the animation on my Linux environment, 1024 bitrate, 1000 frames.
import scipy as sp
import time
from mpl_toolkits.mplot3d import Axes3D
from matplotlib import cm
from matplotlib.ticker import LinearLocator, FormatStrFormatter
import matplotlib.pyplot as plt
import mpl_toolkits.mplot3d.axes3d as p3
import matplotlib.animation as animation
dx=0.01
dy=0.01
a=0.5
timesteps=500
t=0.
nx = int(1/dx)
ny = int(1/dy)
dx2=dx**2
dy2=dy**2
dt = dx2*dy2/( 2*a*(dx2+dy2) )
ui = sp.zeros([nx,ny])
u = sp.zeros([nx,ny])
for i in range(nx):
for j in range(ny):
if ( ( (i*dx-0.5)**2+(j*dy-0.5)**2 <= 0.1)
& ((i*dx-0.5)**2+(j*dy-0.5)**2>=.05) ):
ui[i,j] = 1
def evolve_ts(u, ui):
u[1:-1, 1:-1] = ui[1:-1, 1:-1] + a*dt*(
(ui[2:, 1:-1] - 2*ui[1:-1, 1:-1] + ui[:-2, 1:-1])/dx2 +
(ui[1:-1, 2:] - 2*ui[1:-1, 1:-1] + ui[1:-1, :-2])/dy2 )
def data_gen(framenumber, Z ,surf):
global u
global ui
evolve_ts(u,ui)
ui[:] = u[:]
Z = ui
ax.clear()
plotset()
surf = ax.plot_surface(X, Y, Z, rstride=1, cstride=1, cmap=cm.coolwarm,
linewidth=0, antialiased=False, alpha=0.7)
return surf,
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
X = sp.arange(0,1,dx)
Y = sp.arange(0,1,dy)
X,Y= sp.meshgrid(X,Y)
Z = ui
def plotset():
ax.set_xlim3d(0., 1.)
ax.set_ylim3d(0., 1.)
ax.set_zlim3d(-1.,1.)
ax.set_autoscalez_on(False)
ax.zaxis.set_major_locator(LinearLocator(10))
ax.zaxis.set_major_formatter(FormatStrFormatter('%.02f'))
cset = ax.contour(X, Y, Z, zdir='x', offset=0. , cmap=cm.coolwarm)
cset = ax.contour(X, Y, Z, zdir='y', offset=1. , cmap=cm.coolwarm)
cset = ax.contour(X, Y, Z, zdir='z', offset=-1., cmap=cm.coolwarm)
plotset()
surf = ax.plot_surface(X, Y, Z, rstride=1, cstride=1, cmap=cm.coolwarm,
linewidth=0, antialiased=False, alpha=0.7)
fig.colorbar(surf, shrink=0.5, aspect=5)
ani = animation.FuncAnimation(fig, data_gen, fargs=(Z, surf),frames=1000, interval=30, blit=False)
ani.save("2dDiffusion.mp4", bitrate=1024)
#plt.show()
Tuesday, November 24, 2015
2D Diffusion Equation using Python, Scipy, and VPython
I got it from here, but modify it here and there.
I also add animation using vpython but can't find 3d or surface version, so I planned to go to matplotlib surface plot route, :)
(update: here it is, :) )
.
I also add animation using vpython but can't find 3d or surface version, so I planned to go to matplotlib surface plot route, :)
(update: here it is, :) )
#!/usr/bin/env python
"""
A program which uses an explicit finite difference
scheme to solve the diffusion equation with fixed
boundary values and a given initial value for the
density.
Two steps of the solution are stored: the current
solution, u, and the previous step, ui. At each time-
step, u is calculated from ui. u is moved to ui at the
end of each time-step to move forward in time.
http://www.timteatro.net/2010/10/29/performance-python-solving-the-2d-diffusion-equation-with-numpy/
he uses matplotlib
I use visual python
"""
import scipy as sp
import time
from visual import *
from visual.graph import *
graph1 = gdisplay(x=0, y=0, width=600, height=400,
title='x vs. T', xtitle='x', ytitle='T',
foreground=color.black, background=color.white)
# Declare some variables:
dx=0.01 # Interval size in x-direction.
dy=0.01 # Interval size in y-direction.
a=0.5 # Diffusion constant.
timesteps=500 # Number of time-steps to evolve system.
t=0.
nx = int(1/dx)
ny = int(1/dy)
dx2=dx**2 # To save CPU cycles, we'll compute Delta x^2
dy2=dy**2 # and Delta y^2 only once and store them.
# For stability, this is the largest interval possible
# for the size of the time-step:
dt = dx2*dy2/( 2*a*(dx2+dy2) )
# Start u and ui off as zero matrices:
ui = sp.zeros([nx,ny])
u = sp.zeros([nx,ny])
# Now, set the initial conditions (ui).
for i in range(nx):
for j in range(ny):
if ( ( (i*dx-0.5)**2+(j*dy-0.5)**2 <= 0.1)
& ((i*dx-0.5)**2+(j*dy-0.5)**2>=.05) ):
ui[i,j] = 1
'''
def evolve_ts(u, ui):
global nx, ny
"""
This function uses two plain Python loops to
evaluate the derivatives in the Laplacian, and
calculates u[i,j] based on ui[i,j].
"""
for i in range(1,nx-1):
for j in range(1,ny-1):
uxx = ( ui[i+1,j] - 2*ui[i,j] + ui[i-1, j] )/dx2
uyy = ( ui[i,j+1] - 2*ui[i,j] + ui[i, j-1] )/dy2
u[i,j] = ui[i,j]+dt*a*(uxx+uyy)
'''
def evolve_ts(u, ui):
"""
This function uses a numpy expression to
evaluate the derivatives in the Laplacian, and
calculates u[i,j] based on ui[i,j].
"""
u[1:-1, 1:-1] = ui[1:-1, 1:-1] + a*dt*(
(ui[2:, 1:-1] - 2*ui[1:-1, 1:-1] + ui[:-2, 1:-1])/dx2 +
(ui[1:-1, 2:] - 2*ui[1:-1, 1:-1] + ui[1:-1, :-2])/dy2 )
# Now, start the time evolution calculation...
#tstart = time.time()
f1 = gcurve(color=color.blue)
while True:
rate(60)
#for m in range(1, timesteps+1):
if t<timesteps:
t+=dt
evolve_ts(u, ui)
ui[:] = u[:] # I add this line to update ui value (not present in original code)
#print "Computing u for m =", m
f1.gcurve.pos = []
for i in arange(nx):
f1.plot(pos=(i,u[nx/2,i]))
#tfinish = time.time()
#print "Done."
#print "Total time: ", tfinish-tstart, "s"
#print "Average time per time-step using numpy: ", ( tfinish - tstart )/timesteps, "s."
Monday, November 23, 2015
Numpy Slice Expression
Suppossed we have two array a and b
If we want to set b as finite difference result of a, we may tempted to do this
There's another (faster) way. The performance's close to the pure C, :)
b[:-1] = a[1:]-a[:-1]
What's that?
Numpy has slice form for array. If we have an array with length 10, the a[:] refers to all value in a.
a[1:] refers to a[1] to a[9] (without a[0])
a[3:] refers to a[3] to a[9]
a[:-1] refers to a[0] to a[8]
a[:-3] refers to a[0] to a[6]
a[1:-1] refers to a[1] to a[8]
...
and so on
Here's my tinkering with slice expression
I like Python, :)
If we want to set b as finite difference result of a, we may tempted to do this
for i in range (9):
b[i] = a[i+1]-a[i]
There's another (faster) way. The performance's close to the pure C, :)
b[:-1] = a[1:]-a[:-1]
What's that?
Numpy has slice form for array. If we have an array with length 10, the a[:] refers to all value in a.
a[1:] refers to a[1] to a[9] (without a[0])
a[3:] refers to a[3] to a[9]
a[:-1] refers to a[0] to a[8]
a[:-3] refers to a[0] to a[6]
a[1:-1] refers to a[1] to a[8]
...
and so on
Here's my tinkering with slice expression
>>> from numpy import *
>>> a = zeros(10)
>>> b = zeros(10)
>>> a[5]=1.
>>> a
array([ 0., 0., 0., 0., 0., 1., 0., 0., 0., 0.])
>>> b
array([ 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.])
>>> a[6]=2.
>>> a
array([ 0., 0., 0., 0., 0., 1., 2., 0., 0., 0.])
>>> b[:-1]=a[:-1]-a[1:]
>>> b
array([ 0., 0., 0., 0., -1., -1., 2., 0., 0., 0.])
>>> b[:-1]=a[:-1]+a[1:]
>>> b
array([ 0., 0., 0., 0., 1., 3., 2., 0., 0., 0.])
>>>
Wednesday, November 18, 2015
Create CSV file using Delphi
I used textfile variable to write to a file (or create it if it don't exist).
CSV file? Just make sure that the name at assignfile command had .csv extension, :)
Of course we have to format the output to meet the CSV standart; separated by comma.
.
CSV file? Just make sure that the name at assignfile command had .csv extension, :)
Of course we have to format the output to meet the CSV standart; separated by comma.
procedure TForm1.Button1Click(Sender: TObject);
var
fileku:textfile;
i,j,n:integer;
begin
n:=10;
assignfile(fileku,'data.csv');
rewrite(fileku);
writeln(fileku,'tadaa...');
for i:=1 to n do begin
for j:=1 to n do begin
writeln(fileku,i,',',j,',','data',i,j);
end;
end;
closefile(fileku);
end;
Thursday, November 12, 2015
Delphi on OS X
Here's the WineSkin version.
I found it's way smoother than WineBottler version, ...., but hard to figure how to use it
To install Delphi in OS X using WineSkin, we have to download and install Wineskin, of course, :)
.
I found it's way smoother than WineBottler version, ...., but hard to figure how to use it
To install Delphi in OS X using WineSkin, we have to download and install Wineskin, of course, :)
- Open Wineskin Winery.app
- Make sure you have a Wrapper version and an Engine
- Select the Engine you want to use (I use WS9Wine1.7.52)
- Press the Create Wrapper button
- Enter in the name Delphi (or whatever you have in mind) for the wrapper and press OK
- When its done being created, click the button to view it in Finder in the finished window
- Close Wineskin Winery.app.
- Right click Delphi.app in Finder and select “Show Package Contents”
- Double click and run Wineskin.app.
- Now click on the Install Software button
- Select to choose a setup executable
- Navigate to the Delphi setup exe file you downloaded in step one
- Select the setup exe file and press the choose button
- At this point Delphi setup should begin, go through the Delphi setup like a normal install
- After the setup is done, back in Wineskin.app, it should pop up asking you to select the .exe file
- Choose the delphi32.exe file in the drop down list and press the Select Button
- Now press the Quit button to exit Wineskin.app
- Back in Finder, double click Delphi.app and start coding
code
unit Unit1;
interface
uses
Windows, Messages, SysUtils, Variants, Classes, Graphics, Controls, Forms,
Dialogs, StdCtrls;
type
TForm1 = class(TForm)
Button1: TButton;
procedure Button1Click(Sender: TObject);
procedure FormCreate(Sender: TObject);
private
{ Private declarations }
public
{ Public declarations }
end;
var
Form1: TForm1;
jalan:boolean=false;
implementation
{$R *.dfm}
procedure TForm1.Button1Click(Sender: TObject);
begin
jalan := not jalan;
if jalan = true then button1.Caption:='Stop'
else button1.Caption:='Run';
end;
procedure TForm1.FormCreate(Sender: TObject);
begin
button1.Caption:='Run';
end;
end.
Delphi on OS X
I use WineBottler to install Delphi 7 on my El Capitan.
It's installed, it can run.
The one that tickle me is the toolbar list is scrambled, it's sorted alphabetically, so additional toolbar is in the first and act as default toolbar.
It's installed, it can run.
The one that tickle me is the toolbar list is scrambled, it's sorted alphabetically, so additional toolbar is in the first and act as default toolbar.
code
Sunday, November 8, 2015
SwishMax in Mac.
Subscribe to:
Posts (Atom)
My sky is high, blue, bright and silent.

Nugroho's (almost like junk) blog
By: Nugroho Adi Pramono
323f
(5)
amp
(1)
android
(12)
apple
(7)
arduino
(18)
art
(1)
assembler
(21)
astina
(4)
ATTiny
(23)
blackberry
(4)
camera
(3)
canon
(2)
cerita
(2)
computer
(106)
crazyness
(11)
debian
(1)
delphi
(39)
diary
(286)
flash
(8)
fortran
(6)
freebsd
(6)
google apps script
(8)
guitar
(2)
HTML5
(10)
IFTTT
(7)
Instagram
(7)
internet
(12)
iOS
(5)
iPad
(6)
iPhone
(5)
java
(1)
javascript
(1)
keynote
(2)
LaTeX
(6)
lazarus
(1)
linux
(29)
lion
(15)
mac
(28)
macbook air
(8)
macbook pro
(3)
macOS
(1)
Math
(3)
mathematica
(1)
maverick
(6)
mazda
(4)
microcontroler
(35)
mountain lion
(2)
music
(37)
netbook
(1)
nugnux
(6)
os x
(36)
php
(1)
Physicist
(29)
Picture
(3)
programming
(189)
Python
(109)
S2
(13)
software
(7)
Soliloquy
(125)
Ubuntu
(5)
unix
(4)
Video
(8)
wayang
(3)
yosemite
(3)